Actin filaments are essential to the eukaryotic cell’s cytoskeleton, as they regulate cell shape and allow cells to polarize, protrude, and move.1,2 Evidence was found in the late 1990s that actin holds wave-like dynamics, and filaments propagate within cells.2

Image Credit: Thermo Fisher Scientific
‘Actin waves’ occur in various cells, such as Dictyostelium cells, leukocytes, neurons, osteosarcoma cells, melanoma, oocytes, and embryos. Their function has been proven in many processes, including cell migration, adhesion, cytokinesis, and neurogenesis.2, 3
Until recently, the actin architecture underlying wave propagation was unknown.3
An incomplete picture
Research has demonstrated that actin waves propagate via actin polymerization on the substrate-attached cell membrane at the wave’s front and disassembly at the back of the wave.4 Proteins named Arp2/3 complex stimulate this polymerization, which causes a new filament to branch off from the side of an existing one.
There are two possibilities for how this may occur:
- By the elongation of filaments that point in the wave propagation direction
- By the nucleation of new filaments in front of the wave3
Researchers sought to understand actin wave architecture to establish the real mechanism.
Understanding architecture
Led by Günther Gerisch and Marion Jasnin at the Max Planck Institute of Biochemistry in Martinsried, Germany, researchers utilized in situ cryo-electron tomography (cryo-ET) and cryo-focused ion beam (FIB) sample preparation to study wave propagation in Dictyostelium cells3.
In situ cryo-ET enables high-resolution 3D visualization of proteins in their native cellular environment, while cryo-FIB sample preparation gives access to the cell interior without introducing artifacts.
The team employed these technologies to understand the role played by the Arp2/3 complex in nucleating branches of actin filaments, observing how the direction of the branches correlates with the direction of wave propagation and the orientation of filaments about the substrate-attached cell membrane on which the waves propagate.
The team could not identify a favored alignment of mother or daughter filaments that tallied with wave propagation through analysis of the direction of branch junctions.
This suggested that the mechanism of wave propagation is unlikely to be based on elongating filaments in the direction of the wave propagation, and instead indicated that filament nucleation is more likely to be the wave propagation mechanism.
The researchers calculated the angle between the direction of the substrate-attached cell membrane the mother or daughter filaments, to understand the relation of the geometry of branch nucleation to the membrane on which the waves are propagating.
It was revealed that between 64% and 74% of daughter filaments face the membrane, highlighting that the Arp2/3 complex favors nucleation towards the membrane. However, most mother filaments grow parallel to the membrane.
The researchers discovered that the small portion of mother filaments that are not parallel to the membrane and their daughter filaments build up into tent-like arrays, forming an essential part of establishing the wave architecture.
Through studying live-imaging data, it was demonstrated that VASP, an elongation and nucleation factor, appears to collaborate with the Arp2/3 complex, producing part of the filaments from which the Arp2/3 complex nucleates branches.
A new mechanism
Based on their collective data, the team proposes a mechanism for wave progression in which actin polymerization is stimulated at the membrane by VASP and other elongation and nucleation factors. At these locations of dense actin assembly, the Arp2/3 complex then collects where it nucleates branches.
Filaments grow from these locations towards the membrane and form tent-like arrays. This process repeats, producing new generations of these tent-like arrays as the wave propagates, lifting the prior generations of tent-like arrays with the mother filaments.
“Altogether, the quantitative analysis of branch organization in traveling actin waves reflects the ordered progression in space and time of an actin network from nucleation to depolymerization,” the authors determine in Structure.3
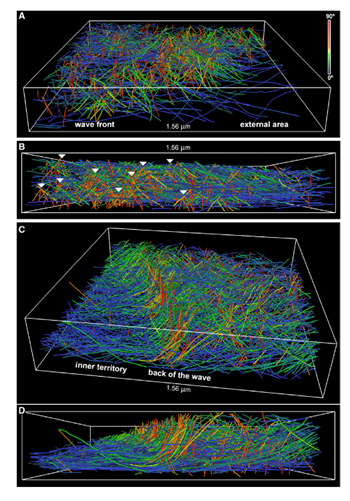
Figure 1. Rendering of actin filaments extracted from tomograms of actin waves showing tent-like arrays. A) View from the front of the wave; B) Side view of the wave; white arrows show tent-like arrays at different distances from the membrane C) Slanting and D) side views of the back of a wave; the tent-like arrays disappear when the wave has passed. Image Credit: Jasnin M et al 2019.
Thermo Fisher Scientific technology
The team employed a Thermo Scientific dual-beam Quanta 3D FIB/SEM, equipped with Thermo Scientific Maps Software.
Thermo Fisher leads the industry in FIB-SEM technology, and this enables researchers to discover subsurface structural detail via the making of specific cuts with a FIB and subsequently imaging the exposed surface with a high-resolution SEM.
Thermo Scientific Maps Software complements this as an imaging and correlative workflow software suite compatible with both Thermo Scientific FIB/SEM and SEM platforms. Maps provides an integrated approach to data acquisition, annotation, and storage, merging correlative microscopy, multi-scale imaging automation, and integrated analytics.
The researchers conducted automated filament segmentation with the use of Thermo ScientificTM AmiraTM Software. This comprised first performing nonlocal-means filtering of the tomograms prior to tracing the actin filaments with an automated segmentation algorithm.
References and further reading
- Merino F, Pospich S, Raunser S. Towards a structural understanding of the remodeling of the actin cytoskeleton. Semin Cell Dev Biol. 2020;102:51-64. doi: 10.1016/j.semcdb.2019.11.018.
- Inagaki N, Katsuno H. Actin Waves: Origin of Cell Polarization and Migration? Trends Cell Biol. 2017;27:515-526. doi: 10.1016/j.tcb.2017.02.003.
- Jasnin M, et al. The Architecture of Traveling Actin Waves Revealed by Cryo-Electron Tomography. Structure. 2019;27(8):1211-1223.e5. doi: 10.1016/j.str.2019.05.009.
- Bretschneider, T, et al. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys. 2009; J 96(7) p2888–2900. doi: 10.1016%2Fj.bpj.2008.12.3942.
Familiarize yourself with Amira Software for Cryo ET
With its advanced image processing, segmentation, quantification, and reporting features powered by artificial intelligence (AI), Amira Software can provide you with the tools you need to take your discoveries to the next level.
In this webinar, you’ll learn how to:
- Reveal and visualize endomembrane system (in situ) and its spatial arrangement within a tomogram
- Quickly reveal and measure the network of filaments involved in cellular processes
- Accelerate image segmentation with tools powered by artificial intelligence (AI)
Watch on-demand Webinar
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.