The advent of genomic editing technologies has revolutionized the fields of microbiology and biotechnology. This article will provide an overview of this subject.
Genome. Image Credit: MIKHAIL GRACHIKOV/Shutterstock.com
Genomic Editing: An Overview
Genes contain the information necessary for coding the proteins in biological cells. Editing genes is the cornerstone of research into the interrogation of the genetic basis of metabolic and physiological processes in living organisms. Over the years, many techniques have been utilized by scientists for this purpose, especially for the investigation of bacteria that have industrial or scientific value, for example, suicide plasmids.
Other approaches for prokaryote genome editing include the lambda Red system and the ClosTron method. Traditional microbial genomic editing techniques have proven to be slow and laborious.
One of the drawbacks of traditional genomic editing methods has been the necessity of introducing resistance marker cassettes into the microbial genome, which is unsuitable for more precise edits such as single amino acid mutations. Several steps are needed including the homologous recombination and excision of unnecessary regions of DNA. Genomic manipulation leaves remnants that interfere with downstream genetic engineering. These drawbacks facilitate the need for better methods.
CRISPR-Cas
CRISPR-Cas is a technology that has dramatically changed the way genomes are edited. This technology has been around for under two decades but has already seen widespread use in multiple fields such as biochemistry, microbiology, drug development, and the food industry. The technique has opened powerful possibilities for tailoring microorganisms for specific purposes.
CRISPR is a technique that is faster, cheaper, and more accurate than conventional DNA editing methods. It enables scientists to edit specific parts of the genome by removing, adding, or altering specific DNA sections.
The system uses two key molecules to introduce a mutation into DNA. These are an enzyme called Cas9, which cuts the double-stranded DNA at a specific location for insertion of a new genetic strand, and guide RNA (gRNA.) gRNA is a pre-designed RNA sequence, usually around 20 base pairs in length contained within a larger RNA “scaffold”, which guides the Cas9 protein to the target DNA sequence for editing. The gRNA has complementary bases to the targeted DNA sequence.
The Cas9 enzyme cuts both strands of DNA and inserts the RNA sequence into the target area of the genetic code. The cell recognizes that the DNA is damaged and repairs it with a new sequence of DNA. Thus, the genetic code of the cell is altered by the CRISPR-Cas9 system and new generations of cells can be made which express the edited genetic code and manufacture new proteins for a multitude of purposes.
The Function of Cas Genes
Cas (CRISPR-associated) genes are commonly found in bacteria and archaea. They act as a way of recognizing foreign DNA and in essence, the CRISPR/Cas system is a primitive immune mechanism. There are multiple Cas genes that have been identified thus far, with Cas9 being the most utilized gene in CRISPR-Cas technologies.
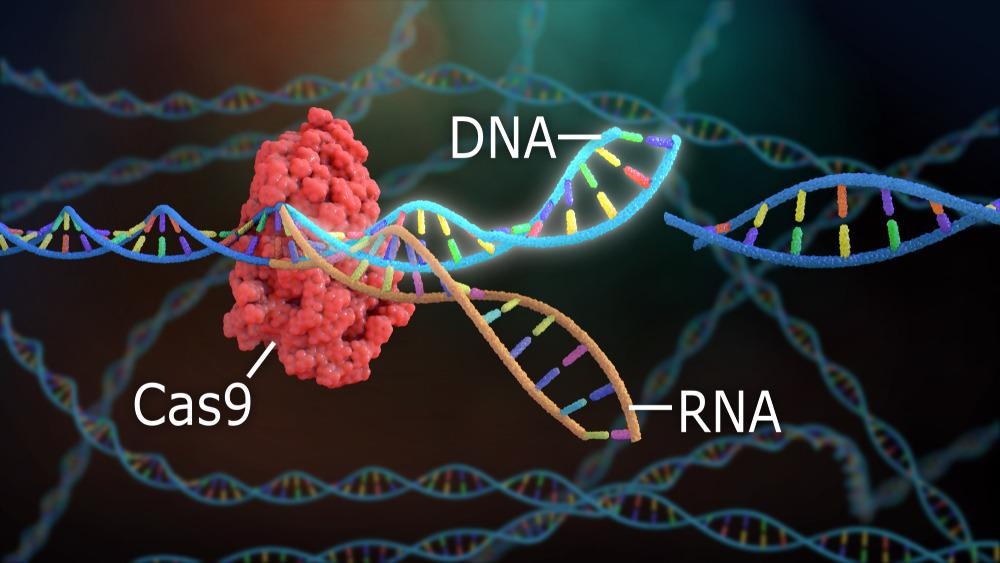
CRISPR-Cas9. Image Credit: Nathan Devery/Shutterstock.com
Microbial Genomic Editing: CRISPR-Cas9 and Beyond
Whilst CRISPR-Cas9 has been widely utilized in editing eukaryotic cells due to its simplicity and cost advantages, the system has been less favored for bacterial genome editing. This is due to challenges that still exist with the technology. A wide array of methods based on CRISPR-Cas9 have been developed to overcome the challenges which vary according to factors such as the number of plasmids used and the specific induced DNA repair mechanism.
One of the main drawbacks to CRISPR-Cas9 methods for genomic editing in microbes is cytotoxicity. This is due to overexpression of Cas9 in the engineered microbes (this is a particular problem with E. coli, but also occurs in other bacterial species) and leads to a lack of colonies. This occurs even when there is no nuclease activity. Additionally, Cas9-mismatch can lead to double-strand breaks at off-target locations within the microbial genome.
Alternative strategies for CRISPR-Cas mediated microbial genome editing have been developed to overcome these drawbacks with the system. These include Scarless Cas9 assisted recombineering, inducing recombinases, encoding DNA repair templates in plasmids, using inducible promoters, nucleases of CRISPR-like DNA Nickase, endogenous CRISPR systems, and using base editors such as adenine deaminase.
Another alternative that has recently been explored is using pre-made ribonucleoprotein (RNP) complexes to deliver CRISPR-Cas machinery instead of using plasmids. This method is highly versatile. An advantage of this method is that it does not rely on the microbial translation and transcription machinery, and direct evaluation of the RNP preparation’s efficacy beforehand via nuclease assays. The RNP complex degrades after use, avoiding overexpression of Cas9.
Summing Up
Microbial genome editing is a powerful way to tailor-make several important products for industries including medicine, food, and industrial chemical production. Many conventional genomic editing methods are laborious, costly, time-consuming, and have a degree of inaccuracy.
CRISPR-Cas has emerged as a central technology in editing the genomes of both eukaryotic and prokaryotic cells, but there are still challenges that prevent its widespread adoption for microbial genomic editing. These challenges are currently being addressed by research into alternative strategies and, though it is still a novel technology, the future of using these methods for engineering microorganisms is promising.
References:
Further Reading